已过期。请访问 使用 Trae 编写 可以携带资产、可以交易的DOBlock - 未编译 未测试 未部署
1. 初始Prompt
在测试网 (Pudge)上,
Cluster的 tx_hash : 0xcebb174d6e300e26074aea2f5dbd7f694bb4fe3de52b6dfe205e54f90164510a
Spore的 tx_hash : 0x5e8d2a517d50fd4bb4d01737a7952a1f1d35c8afc77240695bb569cd7d9d5a1f
一个具体的DOB包括含具体id的cluster和含具体id的spore
请编写一个DOBLock,UTXO上的CKB只能由某个DOB解锁
#![no_std]
#![no_main]
#![feature(lang_items)]
#![feature(alloc_error_handler)]
#![feature(panic_info_message)]
// Import CKB syscalls and high-level APIs
use ckb_std::{
ckb_constants::Source,
ckb_types::prelude::*,
default_alloc,
entry,
error::SysError,
high_level::{load_script, load_cell_data, load_cell_type, load_cell_type_hash, QueryIter},
};
// Constants that you need to define based on your Spore protocol deployment
// Example: Replace with actual Spore Type Script Code Hash and Hash Type
const SPORE_TYPE_CODE_HASH: [u8; 32] = [0x00, /* ... 30 more bytes ... */ 0x00];
const SPORE_TYPE_HASH_TYPE: u8 = 0x01; // 0x00 for data, 0x01 for type, 0x02 for data1
// Define the expected length of Cluster ID and Spore ID
// Adjust these if your IDs have different lengths
const CLUSTER_ID_LEN: usize = 32;
const SPORE_ID_LEN: usize = 32;
// Error codes
mod error {
use ckb_std::error::SysError;
// See: https://github.com/nervosnetwork/ckb-system-scripts/blob/master/c/error.h
// And: https://github.com/nervosnetwork/ckb-vm/blob/master/versions/VM1/asm/riscv_asm.h
pub const ERROR_ARGS_LEN: i8 = -1;
pub const ERROR_ENCODING: i8 = -2;
pub const ERROR_SYSCALL: i8 = -3;
pub const ERROR_SCRIPT_TOO_LONG: i8 = -21;
pub const ERROR_VALIDATION_FAILURE: i8 = -31;
pub const ERROR_INVALID_MOL_FORMAT: i8 = -41;
pub const ERROR_DOB_NOT_FOUND_IN_INPUTS: i8 = 50;
pub const ERROR_CLUSTER_ID_MISMATCH: i8 = 51;
pub const ERROR_SPORE_ID_MISMATCH: i8 = 52;
pub const ERROR_INVALID_SPORE_CELL_DATA: i8 = 53;
}
// Main entry point
fn main() -> i8 {
// 1. Load current script (the DOBLock)
let script = match load_script() {
Ok(s) => s,
Err(e) => return e.into(),
};
let args: &[u8] = script.args().raw_data();
// 2. Validate arguments length
// Expected: target_cluster_id (32 bytes) + target_spore_id (32 bytes)
if args.len() != CLUSTER_ID_LEN + SPORE_ID_LEN {
return error::ERROR_ARGS_LEN;
}
let target_cluster_id = &args[0..CLUSTER_ID_LEN];
let target_spore_id = &args[CLUSTER_ID_LEN..(CLUSTER_ID_LEN + SPORE_ID_LEN)];
// 3. Iterate through transaction inputs to find the required DOB
let mut dob_found = false;
for i in 0.. {
// Load cell type script from input
match load_cell_type(i, Source::Input) {
Ok(Some(input_type_script)) => {
// Check if this is a Spore Cell
if input_type_script.code_hash().raw_data().as_ref() == SPORE_TYPE_CODE_HASH
&& input_type_script.hash_type().as_slice() == [SPORE_TYPE_HASH_TYPE]
{
let input_spore_id = input_type_script.args().raw_data();
// Check Spore ID
if input_spore_id.as_ref() != target_spore_id {
// Not the target Spore, continue searching
continue;
}
// Spore ID matches, now check Cluster ID from cell data
match load_cell_data(i, Source::Input) {
Ok(spore_cell_data) => {
// TODO: Implement the logic to parse cluster_id from spore_cell_data
// This depends on how cluster_id is stored in your Spore's data structure.
// For example, if cluster_id is the first 32 bytes of the data:
if spore_cell_data.len() >= CLUSTER_ID_LEN {
let input_cluster_id = &spore_cell_data[0..CLUSTER_ID_LEN];
if input_cluster_id == target_cluster_id {
dob_found = true;
break; // DOB found, exit loop
} else {
// Cluster ID mismatch for the target Spore ID
// This might be an error condition depending on your exact requirements
// For now, we'll assume it's just not the DOB we're looking for if cluster_id doesn't match.
// If a Spore ID can only belong to one Cluster, then this is an error.
// return error::ERROR_CLUSTER_ID_MISMATCH;
}
} else {
// Spore cell data is too short to contain a cluster_id
return error::ERROR_INVALID_SPORE_CELL_DATA;
}
}
Err(SysError::IndexOutOfBound) => {
// Should not happen if load_cell_type was successful
break;
}
Err(_) => return error::ERROR_SYSCALL, // Or a more specific error
}
}
}
Ok(None) => {
// No type script for this input, skip
continue;
}
Err(SysError::IndexOutOfBound) => {
// Reached the end of inputs
break;
}
Err(e) => return e.into(), // Other syscall error
}
}
if dob_found {
0 // Success
} else {
error::ERROR_DOB_NOT_FOUND_IN_INPUTS
}
}
// Default allocators and panic handlers for no_std environment
#[global_allocator]
static ALLOC: default_alloc::DefaultAllocator = default_alloc::DefaultAllocator;
#[alloc_error_handler]
fn oom_handler(_layout: core::alloc::Layout) -> ! {
panic!("Out of memory");
}
#[panic_handler]
fn panic_handler(info: &core::panic::PanicInfo) -> ! {
if let Some(s) = info.message() {
ckb_std::syscalls::debug(alloc::format!("panic: {:?}", s).as_str());
} else {
ckb_std::syscalls::debug("panic occurred");
}
ckb_std::syscalls::exit(-1);
}
#[lang = "eh_personality"]
extern "C" fn eh_personality() {}
entry!(main);
2. 通用化,不硬编码code_hash
如果想让这个lock更加通用,适用于任意版本的Spore,就不应该硬编码cluster和spore的code_hash,应该把它们放在args或其它字段中,应该怎么更新代码?
#![no_std]
#![no_main]
#![feature(lang_items)]
#![feature(alloc_error_handler)]
#![feature(panic_info_message)]
// Import CKB syscalls and high-level APIs
use ckb_std::{
ckb_constants::Source,
ckb_types::prelude::*,
default_alloc,
entry,
error::SysError,
high_level::{load_script, load_cell_data, load_cell_type, QueryIter},
};
// Define the expected length of Cluster ID and Spore ID
// Adjust these if your IDs have different lengths
const CODE_HASH_LEN: usize = 32;
const HASH_TYPE_LEN: usize = 1;
const CLUSTER_ID_LEN: usize = 32;
const SPORE_ID_LEN: usize = 32;
// Error codes
mod error {
// ... existing code ...
pub const ERROR_ARGS_LEN: i8 = -1;
pub const ERROR_ENCODING: i8 = -2;
pub const ERROR_SYSCALL: i8 = -3;
pub const ERROR_SCRIPT_TOO_LONG: i8 = -21;
pub const ERROR_VALIDATION_FAILURE: i8 = -31;
pub const ERROR_INVALID_MOL_FORMAT: i8 = -41;
pub const ERROR_DOB_NOT_FOUND_IN_INPUTS: i8 = 50;
pub const ERROR_CLUSTER_ID_MISMATCH: i8 = 51;
pub const ERROR_SPORE_ID_MISMATCH: i8 = 52;
pub const ERROR_INVALID_SPORE_CELL_DATA: i8 = 53;
}
// Main entry point
fn main() -> i8 {
// 1. Load current script (the DOBLock)
let script = match load_script() {
Ok(s) => s,
Err(e) => return e.into(),
};
let args: &[u8] = script.args().raw_data();
// 2. Validate arguments length
// Expected: spore_code_hash (32) + spore_hash_type (1) + cluster_id (32) + spore_id (32)
const EXPECTED_ARGS_LEN: usize = CODE_HASH_LEN + HASH_TYPE_LEN + CLUSTER_ID_LEN + SPORE_ID_LEN;
if args.len() != EXPECTED_ARGS_LEN {
return error::ERROR_ARGS_LEN;
}
let mut offset = 0;
let target_spore_type_code_hash = &args[offset..offset + CODE_HASH_LEN];
offset += CODE_HASH_LEN;
let target_spore_type_hash_type = args[offset]; // Read the single byte for hash_type
offset += HASH_TYPE_LEN;
let target_cluster_id = &args[offset..offset + CLUSTER_ID_LEN];
offset += CLUSTER_ID_LEN;
let target_spore_id = &args[offset..offset + SPORE_ID_LEN];
// 3. Iterate through transaction inputs to find the required DOB
let mut dob_found = false;
for i in 0.. {
// Load cell type script from input
match load_cell_type(i, Source::Input) {
Ok(Some(input_type_script)) => {
// Check if this is the target Spore Cell type
if input_type_script.code_hash().raw_data().as_ref() == target_spore_type_code_hash
&& input_type_script.hash_type().as_slice() == [target_spore_type_hash_type]
{
let input_spore_id = input_type_script.args().raw_data();
// Check Spore ID
if input_spore_id.as_ref() != target_spore_id {
// Not the target Spore, continue searching
continue;
}
// Spore ID matches, now check Cluster ID from cell data
match load_cell_data(i, Source::Input) {
Ok(spore_cell_data) => {
// TODO: Implement the logic to parse cluster_id from spore_cell_data
// This depends on how cluster_id is stored in your Spore's data structure.
// For example, if cluster_id is the first CLUSTER_ID_LEN bytes of the data:
if spore_cell_data.len() >= CLUSTER_ID_LEN {
let input_cluster_id = &spore_cell_data[0..CLUSTER_ID_LEN];
if input_cluster_id == target_cluster_id {
dob_found = true;
break; // DOB found, exit loop
} else {
// Cluster ID mismatch for the target Spore ID
// return error::ERROR_CLUSTER_ID_MISMATCH;
}
} else {
// Spore cell data is too short to contain a cluster_id
return error::ERROR_INVALID_SPORE_CELL_DATA;
}
}
Err(SysError::IndexOutOfBound) => {
// Should not happen if load_cell_type was successful
break;
}
Err(_) => return error::ERROR_SYSCALL, // Or a more specific error
}
}
}
Ok(None) => {
// No type script for this input, skip
continue;
}
Err(SysError::IndexOutOfBound) => {
// Reached the end of inputs
break;
}
Err(e) => return e.into(), // Other syscall error
}
}
if dob_found {
0 // Success
} else {
error::ERROR_DOB_NOT_FOUND_IN_INPUTS
}
}
// ... existing allocator and panic handler code ...
#[global_allocator]
static ALLOC: default_alloc::DefaultAllocator = default_alloc::DefaultAllocator;
#[alloc_error_handler]
fn oom_handler(_layout: core::alloc::Layout) -> ! {
panic!("Out of memory");
}
#[panic_handler]
fn panic_handler(info: &core::panic::PanicInfo) -> ! {
if let Some(s) = info.message() {
ckb_std::syscalls::debug(alloc::format!("panic: {:?}", s).as_str());
} else {
ckb_std::syscalls::debug("panic occurred");
}
ckb_std::syscalls::exit(-1);
}
#[lang = "eh_personality"]
extern "C" fn eh_personality() {}
entry!(main);
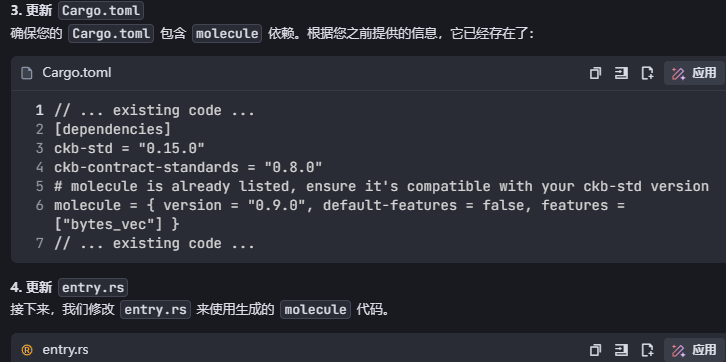
3. 引入molecule
Nervos官方有一个负责序列化和反序列化的库 molecule,请引入它处理args中的数据,以便让它更容易阅读
// Import CKB standard library
use ckb_std::{
ckb_constants::Source,
ckb_types::core::ScriptHashType,
ckb_types::{bytes::Bytes, prelude::*},
high_level::*,
};
// Import the generated molecule code
// Assuming you named the generated file dob_lock_args_mol.rs and placed it in src
mod dob_lock_args_mol;
use dob_lock_args_mol::DOBLockArgs;
// Import Spore/Cluster related constants or functions if needed
// For example, if Spore ID is stored in a specific part of the data
// or if Cluster ID needs specific parsing from Spore data.
// const SPORE_TYPE_SCRIPT_ARGS_CLUSTER_ID_OFFSET: usize = 32; // Example offset for Cluster ID in Spore type args
// const SPORE_TYPE_SCRIPT_ARGS_CLUSTER_ID_LEN: usize = 32; // Example length for Cluster ID
// Define error codes
#[repr(i8)]
enum Error {
IndexOutOfBound = 1,
ItemMissing,
LengthNotEnough,
Encoding,
// Add more error codes as needed
ArgsInvalid,
SporeCellNotFound,
SporeTypeMismatch,
ClusterIdMismatch,
SporeIdMismatch,
InvalidHashType,
}
impl From<molecule::error::VerificationError> for Error {
fn from(_: molecule::error::VerificationError) -> Self {
Error::Encoding
}
}
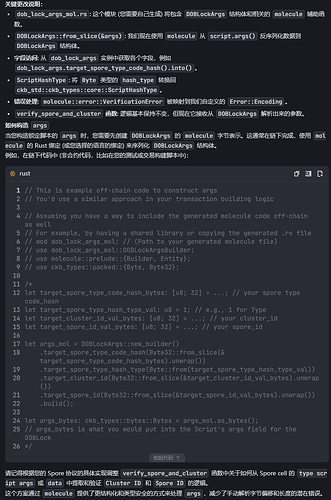
fn verify_spore_and_cluster(
target_spore_type_code_hash: &[u8; 32],
target_spore_type_hash_type: ScriptHashType,
target_cluster_id_bytes: &[u8; 32],
target_spore_id_bytes: &[u8; 32],
) -> Result<(), Error> {
let mut spore_found = false;
// Iterate through inputs to find the Spore cell
for i in 0.. {
match load_cell_type(i, Source::Input) {
Ok(Some(script)) => {
// 1. Check if the type script code_hash and hash_type match the target Spore type
if script.code_hash().raw_data().as_ref() == target_spore_type_code_hash
&& script.hash_type().into() == target_spore_type_hash_type
{
// This input cell has the target Spore type script.
// Now, we need to verify Cluster ID and Spore ID.
// 2. Verify Cluster ID
// Cluster ID is typically part of the Spore's type script args or its data.
// This part depends heavily on how your Spore protocol defines Cluster ID association.
// For this example, let's assume Cluster ID is the first 32 bytes of the Spore's type script args.
// Adjust this logic based on your actual Spore protocol.
let spore_type_args: Bytes = script.args().unpack();
if spore_type_args.len() < 32 { // Assuming Cluster ID is at least 32 bytes
continue; // Not enough args for Cluster ID, try next input
}
let cluster_id_from_spore_args = &spore_type_args[0..32];
if cluster_id_from_spore_args != target_cluster_id_bytes {
continue; // Cluster ID mismatch, try next input
}
// 3. Verify Spore ID
// Spore ID is typically the first 32 bytes of the Spore cell's data.
// Adjust this logic based on your actual Spore protocol.
let spore_data = load_cell_data(i, Source::Input)?;
if spore_data.len() < 32 { // Assuming Spore ID is at least 32 bytes
continue; // Not enough data for Spore ID, try next input
}
let spore_id_from_data = &spore_data[0..32];
if spore_id_from_data != target_spore_id_bytes {
continue; // Spore ID mismatch, try next input
}
// If all checks pass, we found the required Spore
spore_found = true;
break;
}
}
Ok(None) => {
// No type script for this input cell
}
Err(Error::IndexOutOfBound) => {
break; // No more input cells
}
Err(e) => return Err(e), // Other errors
}
}
if spore_found {
Ok(())
} else {
Err(Error::SporeCellNotFound) // Or a more specific error like ClusterIdMismatch or SporeIdMismatch
}
}
pub fn main() -> Result<(), Error> {
// Load the current script (DOBLock)
let script = load_script()?;
let args: Bytes = script.args().unpack();
// Deserialize args using Molecule
let dob_lock_args = DOBLockArgs::from_slice(&args).map_err(|_| Error::ArgsInvalid)?;
let target_spore_type_code_hash: [u8; 32] = dob_lock_args.target_spore_type_code_hash().into();
let target_spore_type_hash_type_byte: u8 = dob_lock_args.target_spore_type_hash_type().into();
let target_cluster_id_bytes: [u8; 32] = dob_lock_args.target_cluster_id().into();
let target_spore_id_bytes: [u8; 32] = dob_lock_args.target_spore_id().into();
let target_spore_type_hash_type = match target_spore_type_hash_type_byte {
0 => ScriptHashType::Data,
1 => ScriptHashType::Type,
2 => ScriptHashType::Data1,
4 => ScriptHashType::Data2, // CKB2023
_ => return Err(Error::InvalidHashType),
};
// The core logic: verify that a Spore cell matching the IDs is present in the inputs.
// This part of the logic assumes that the presence of the specific Spore cell in the inputs
// is sufficient to unlock the CKB. If other conditions are needed (e.g., witness arguments),
// they should be added here.
verify_spore_and_cluster(
&target_spore_type_code_hash,
target_spore_type_hash_type,
&target_cluster_id_bytes,
&target_spore_id_bytes,
)?;
Ok(())
}